| |
| Names | |
|---|---|
| IUPAC name
1-Hydroxypropane-1,2,3-tricarboxylic acid
| |
| Identifiers | |
3D model (JSmol)
|
|
| ChEBI | |
| ChEMBL | |
| ChemSpider | |
| DrugBank |
|
| ECHA InfoCard | 100.005.713 |
| KEGG | |
| MeSH | Isocitrate |
PubChem CID
|
|
| UNII | |
CompTox Dashboard (EPA)
|
|
| |
| |
| Properties | |
| C6H8O7 | |
| Molar mass | 192.124 |
| Melting point | 105 °C (221 °F; 378 K) |
Except where otherwise noted, data are given for materials in their standard state (at 25 °C [77 °F], 100 kPa).
| |
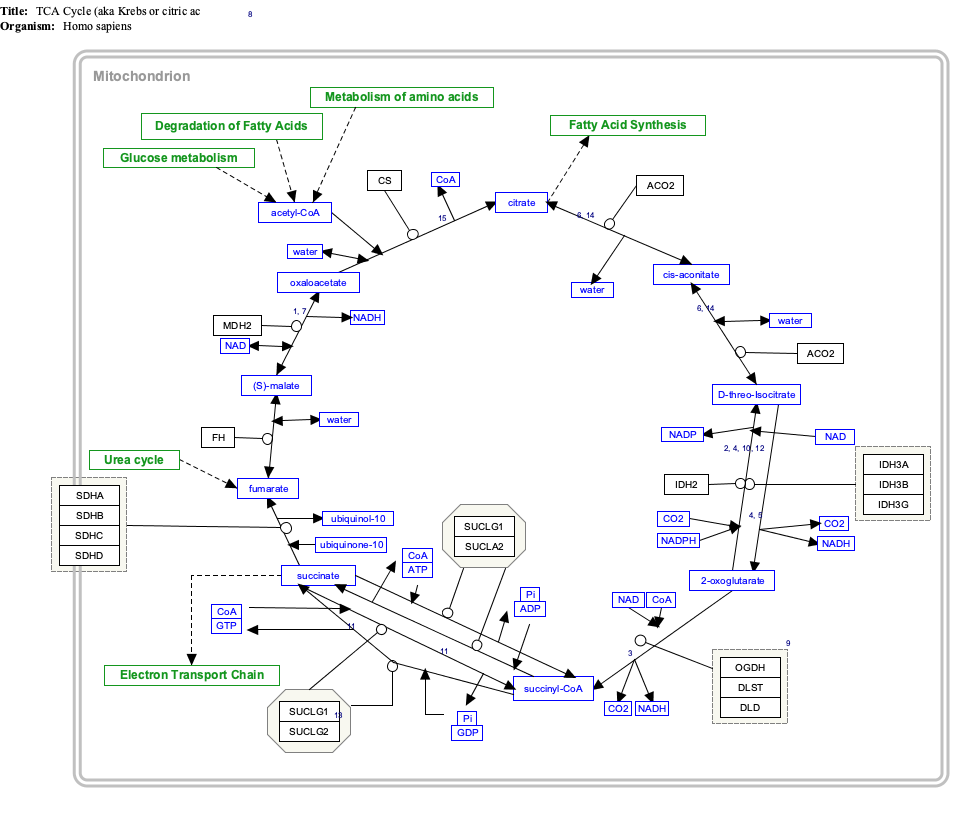
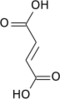
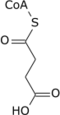
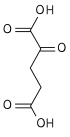
Isocitric acid is a structural isomer of citric acid. Since citric acid and isocitric acid are structural isomers, they share similar physical and chemical properties. Due to these similar properties, it is difficult to separate the isomers.[1] Salts and esters of isocitric acid are known as isocitrates. The isocitrate anion is a substrate of the citric acid cycle. Isocitrate is formed from citrate with the help of the enzyme aconitase, and is acted upon by isocitrate dehydrogenase.
Isocitric acid is commonly used as a marker to detect the authenticity and quality of fruit products, most often citrus juices. In authentic orange juice, for example, the ratio of citric acid to D-isocitric acid is usually less than 130. An isocitric acid value higher than this may be indicative of fruit juice adulteration.[2]
Isocitric acid has largely been used as a biochemical agent due to limited amounts.[3] However, isocitric acid has been shown to have pharmaceutical and therapeutic effects. Isocitric acid has been shown to effectively treat iron deficient anemia.[1] Additionally, isocitric acid could be used to treat Parkinson's disease.[3] Yarrowia lipolytica can be used to produce isocitric acid and is inexpensive compared to other methods. Furthermore, other methods produce unequal amounts of citric acid to isocitric acid ratio, mostly producing citric acid. Use of Yarrowia lipolytica produces a better yield, making equal amounts of citric acid to isocitric acid.[3]
YouTube Encyclopedic
-
1/5Views:32 26110 6891 6503 976432
-
Citric Acid Cycle Mnemonic (B: 2/5): Biochem Tutorial: Krebs cycle, substrates, citrate, isocitrate
-
28. Kevin Ahern's Biochemistry - Citric Acid Cycle II
-
MECHANISM OF AEROBIC RESPIRATION_PART 02
-
Κύκλος Κιτρικού Οξέος (Krebs citric acid cycle) - DigiClass
-
Magic plant House leek
Transcription
Our previous tutorials discussed glycolysis and the transition from pyruvate to the citric acid cycle. The substrates of the citric krebs cycle can be remembered by this mnemonic: Access is the only key to suck a sucky few more orphan cities or Access “is C” only “key to” |suck A| “suck E” few more orphan cities. Access means “cis-Aconitate” “is C” means “IsoCitrate” “only” means “Oxalosuccinate” “key to” means “a-Ketoglutarate” “suck A” means “Succinyl-CoA” “sucky” means Succinate “Few” means “Fumarate” “More” means “malate” “Orphan” means “Oxaloacetate” “Cities” means “Citrate” This mnemonic can loop endlessley, just like the citric acid cycle Access is the only key to suck a “sucky” few more orphan city access is the only key to suck a sucky few more orphan cities. Our next biochemistry tutorial explains the enyzmes involved, and how you can guess their names from the nearby substrates. HelpHippo: Help your Hippocampus
Interactive pathway map
Click on genes, proteins and metabolites below to link to respective articles. [§ 1]
- ^ The interactive pathway map can be edited at WikiPathways: "TCACycle_WP78".
See also
References
- ^ a b Kamzolova, Svetlana V.; Morgunov, Igor G. (2019-12-01). "Microbial production of (2R,3S)-isocitric acid: state of the arts and prospects". Applied Microbiology and Biotechnology. 103 (23): 9321–9333. doi:10.1007/s00253-019-10207-4. ISSN 1432-0614. PMID 31748825.
- ^ Saavedra, L.; Garcia, A.; Barbas, C. (9 June 2000). "Development and validation of a capillary electrophoresis method for direct measurement of isocitric, citric, tartaric and malic acids as adulteration markers in orange juice". Journal of Chromatography A. 881 (1–2): 395–401. doi:10.1016/s0021-9673(00)00258-2. PMID 10905722.
- ^ a b c Yuzbasheva, Evgeniya Y.; Scarcia, Pasquale; Yuzbashev, Tigran V.; Messina, Eugenia; Kosikhina, Iuliia M.; Palmieri, Luigi; Shutov, Artem V.; Taratynova, Maria O.; Amaro, Rodrigo Ledesma; Palmieri, Ferdinando; Sineoky, Sergey P. (2021-05-01). "Engineering Yarrowia lipolytica for the selective and high-level production of isocitric acid through manipulation of mitochondrial dicarboxylate–tricarboxylate carriers". Metabolic Engineering. 65: 156–166. doi:10.1016/j.ymben.2020.11.001. ISSN 1096-7176. PMID 33161142. S2CID 226286865.