| EPSP Synthase (3-phosphoshikimate 1-carboxyvinyltransferase) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
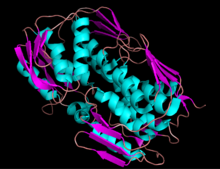
 EPSP synthase liganded with shikimate.[1] | |||||||||
| Identifiers | |||||||||
| EC no. | 2.5.1.19 | ||||||||
| CAS no. | 9068-73-9 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / QuickGO | ||||||||
| |||||||||
5-enolpyruvylshikimate-3-phosphate (EPSP) synthase is an enzyme produced by plants and microorganisms. EPSPS catalyzes the chemical reaction:
- phosphoenolpyruvate (PEP) + 3-phospho shikimate (S3P) ⇌ phosphate + 5-enolpyruvylshikimate-3-phosphate (EPSP)
Thus, the two substrates of this enzyme are phosphoenolpyruvate (PEP) and 3-phosphoshikimate, whereas its two products are phosphate and 5-enolpyruvylshikimate-3-phosphate.
This enzyme is not present in the genomes of animals. It presents an attractive biological target for herbicides, such as glyphosate. A glyphosate-resistant version of this gene has been used in genetically modified crops.
YouTube Encyclopedic
-
1/5Views:6 5159686 19122 16851 039
-
Inhibition by Roundup
-
Amino Acid Synthesis Inhibitors
-
El estado nutricional de los Transgénicos por el Dr. THIERRY VRAIN
-
Stepping through the Shikimate Pathway I
-
Pharma Tube - 37 - CNS - 1 - Brain Subdivision and Central Neurotransmitters [HD]
Transcription
Remembering that phosphorylation is nature's way of transforming a hydroxyl group into a good leaving group, you can find a couple of examples of this strategy in the shikimate pathway. I won't spend much time on this one. I've already talked about it just a little bit, but the chorismate synthase is this 1,4 addition where the phosphate group is lost to introduce a new degree of unsaturation into the six-membered ring. The second, and much more interesting example, is the substitution reaction that's shown here. So we're going to do a substitution reaction on the enol, phosphoenol- pyruvate sp2 carbon, so we're going to take and at that sp2 center, replace the phosphate group with the hydroxyl group on this shikimate acid. And so how does that take place? Well, that mec- mechanism as we'll see takes place by an additional elimination process. Actually, you should by now be used to thinking about substitution at sp2 centers as taking place by this mechanism, there'll be a key tetrahedral intermediate that gets involved. It's all catalyzed by EPSP synthase, enolpyruvate shikimate phosphate synthase, and this is the step where the inhibitor that's in Roundup gets involved and basically blocks the, ah, this addition elimination, ah, mechanism from taking place. So let's understand the mechanism so we can understand how Roundup, a competitive inhibitor, that's a transition state analog of a key portion of the rate- the rate determining step of this, ah, substitution reaction - how does it do it's interference, how does it cause it's, ah, interference? Well, it actually is all in the first step that I'm going to show you where, as we're going to get set up to do that addition elimination, the first thing that happens is this double bond of the phosphoenolpyruvate, which benefits from donation of electron density from the nonbonding pair of electrons into π*, makes that a fairly nucleophilic carbon-carbon double bond. And, and so, the enzyme active site has a side chain that delivers a proton to that, and we can think about this step as an electrophile association step, and in fact, that's how we want to understand it in terms of trying to understand how the inhibitor Roundup gets involved. This is in fact the rate determining step for the overall substitution process. And if I draw it as this, without first invoking that lone pair, but I just think about this as an electrophile association step, which of course as you know will make a carbocation, we see that the structure of the, ah, intermediate is going to have a partial positive charge at the position, ah, that, that's shown there. That's key because we want to mimic that in our transition state analog, the phosph-, the gly- glyphos that's, ah, the inhibitor that is present in Roundup. Well, there's obviously another important resonance contributor, that's the, ah, N to A donation that we see there that puts the, ah, carbon- oxygen double bond that's going to be a very good electrophile for the first step, or the second step of this process. This is actually the addition step, so up till now, we've just been getting ready to do the addition step of the addition elimination substitution reaction. So let's see that, how that unfolds. Here we have, ah, the shikimate acid and it's going to be this hydroxyl group, which is going to serve as the nucleophile. It's going to be general base-catalyzed, and, and so the addition step looks like this. It's a general-base catalyzed ADN step that's going to make a new oxygen, ah, carbon bond and the structure of that is shown over here. We've now protonated the conjugate, ah, the base, as, as its conjugate acid and so it can serve as a general acid in doing an elimination step. So, our, ah, what was functioning as the acid is now positioned as a base and that's going to be used to eliminate one of the carbon- hydrogen bonds, we'll kick out the good leaving group that's going to be general acid catalyzed, and so the final substitution reaction, which we needed to return that carbon back to its sp2 form, that ah where the substitution reaction takes place, gives us that structure. Alright, now we want to understand how, ah, the inhibitor glyphos mimics the rate determining step, which is the generation of this intermediate that is set up to do the addition process. So here's the overall step. Basically, it's the developing positive charge here, and we can see that in the transition state, um, there's, ah, that is, um, in this, for the addition step that there's a partial positive charge on that carbon. It has a geometry that's beginning to take a tetrahedral geometry and that's the, that is what we're trying to mimic in the inhibitor. We want to fit a structure in that will basically be completely stable, but block the active site by mimicking the electronic structure, the partial positive charge, and the geometry as being tetrahedral. And that's the enzyme inhibitor; phosphonomethyl glycine is the formal name. Ah, most people who use this call it glyphos. It's a carbocation mimic. It's, ah, basically going to have the geometry and electronic features of this, ah, transition state structure here. We can see that by doing our superposition idea, and you can see how phosphonomethyl glycine overlays very nicely on top of this transition structure that we drew. I was, ah, I grew up on a farm and I spent, ah, a, many summers walking soy beans because they needed to be weeded. Um, sometime in the early 1990s there was the introduction of a new breed of soybeans, Roundup ready soybeans. This is a wonderful marriage of genetics and molecular biology with chemistry because this was a genetic mutant of the EPSP synthase enzyme which was tolerant to glycine, ah, this ah, to, to, to the, the phosphonomethyl glycine and, ah, and so it's really a remarkable feat in that we have soybeans that basically have a mutated enzyme which can um basically survive in the presence of Roundup so that one can spray the soybean field and you now see these spectacularly clean soybean fields today. People don't have to do the suffering of the summer heat that I had to experience when I was walking soybeans as a, as a, ah, as a teenager.
Nomenclature
The enzyme belongs to the family of transferases, to be specific those transferring aryl or alkyl groups other than methyl groups. The systematic name of this enzyme class is phosphoenolpyruvate:3-phosphoshikimate 5-O-(1-carboxyvinyl)-transferase. Other names in common use include:
- 5-enolpyruvylshikimate-3-phosphate synthase,
- 3-enolpyruvylshikimate 5-phosphate synthase,
- 3-enolpyruvylshikimic acid-5-phosphate synthetase,
- 5′-enolpyruvylshikimate-3-phosphate synthase,
- 5-enolpyruvyl-3-phosphoshikimate synthase,
- 5-enolpyruvylshikimate-3-phosphate synthetase,
- 5-enolpyruvylshikimate-3-phosphoric acid synthase,
- enolpyruvylshikimate phosphate synthase, and
- 3-phosphoshikimate 1-carboxyvinyl transferase.
Structure
EPSP synthase is a monomeric enzyme with a molecular mass of about 46,000.[2][3][4] It is composed of two domains, which are joined by protein strands. This strand acts as a hinge, and can bring the two protein domains closer together. When a substrate binds to the enzyme, ligand bonding causes the two parts of the enzyme to clamp down around the substrate in the active site.
EPSP synthase has been divided into two groups according to glyphosate sensitivity. Class I enzyme, contained in plants and in some bacteria, is inhibited at low micromolar glyphosate concentrations, whereas class II enzyme, found in other bacteria, is resistant to inhibition by glyphosate.[5]
Shikimate pathway
EPSP synthase participates in the biosynthesis of the aromatic amino acids phenylalanine, tyrosine, and tryptophan via the shikimate pathway in bacteria, fungi, and plants. EPSP synthase is produced only by plants and micro-organisms; the gene coding for it is not in the mammalian genome.[6][7] Gut flora of some animals contain EPSPS.[8]
Reaction
EPSP synthase catalyzes the reaction which converts shikimate-3-phosphate plus phosphoenolpyruvate to 5-enolpyruvylshikimate-3-phosphate (EPSP) by way of an acetal-like tetrahedral intermediate.[9][10] Basic and amino acids in the active site are involved in deprotonation of the hydroxyl group of PEP and in the proton-exchange steps related to the tetrahedral intermediate itself, respectively.[11]
Studies of the enzyme kinetics for this reaction have determined the specific sequence and energetics of each step of the process.[12] A deprotonated lysine22 acts as a general base, deprotonating the hydroxyl of S3P such that the resulting oxyanion can attack the most electrophilic carbon of PEP. Glutamate341 acts as a general acid by donating a H+. The deprotonated glutamate341 then acts as a base, taking back its proton, and the S3P group is kicked off and protonated by the protonated lysine.
Herbicide target
EPSP synthase is the biological target for the herbicide glyphosate.[13] Glyphosate is a competitive inhibitor of EPSP synthase, acting as a transition state analog that binds more tightly to the EPSPS-S3P complex than PEP and inhibits the shikimate pathway. This binding leads to inhibition of the enzyme's catalysis and shuts down the pathway. Eventually this results in organism death from lack of aromatic amino acids the organism requires to survive.[5][14]
A version of the enzyme that both was resistant to glyphosate and that was still efficient enough to drive adequate plant growth was identified by Monsanto scientists after much trial and error in an Agrobacterium strain called CP4 (Q9R4E4). The strain CP4 was found surviving in a waste-fed column at a glyphosate production facility. The CP4 EPSP synthase enzyme has been engineered into several genetically modified crops.[5][15]
References
- ^ Priestman MA, Healy ML, Funke T, Becker A, Schönbrunn E (October 2005). "Molecular basis for the glyphosate-insensitivity of the reaction of 5-enolpyruvylshikimate 3-phosphate synthase with shikimate". FEBS Lett. 579 (25): 5773–80. doi:10.1016/j.febslet.2005.09.066. PMID 16225867. S2CID 26614581.
- ^ Goldsbrough, Peter (1990). "Gene amplification in glyphosate tolerant tobacco cells". Plant Science. 72 (1): 53–62. doi:10.1016/0168-9452(90)90186-r.
- ^ Abdel-Meguid SS, Smith WW, Bild GS (Dec 1985). "Crystallization of 5-enolpyruvylshikimate 3-phosphate synthase from Escherichia coli". Journal of Molecular Biology. 186 (3): 673. doi:10.1016/0022-2836(85)90140-8. PMID 3912512.
- ^ Ream JE, Steinrücken HC, Porter CA, Sikorski JA (May 1988). "Purification and Properties of 5-Enolpyruvylshikimate-3-Phosphate Synthase from Dark-Grown Seedlings of Sorghum bicolor". Plant Physiology. 87 (1): 232–8. doi:10.1104/pp.87.1.232. PMC 1054731. PMID 16666109.
- ^ a b c Pollegioni L, Schonbrunn E, Siehl D (Aug 2011). "Molecular basis of glyphosate resistance-different approaches through protein engineering". The FEBS Journal. 278 (16): 2753–66. doi:10.1111/j.1742-4658.2011.08214.x. PMC 3145815. PMID 21668647.
- ^ Funke T, Han H, Healy-Fried ML, Fischer M, Schönbrunn E (Aug 2006). "Molecular basis for the herbicide resistance of Roundup Ready crops". Proceedings of the National Academy of Sciences of the United States of America. 103 (35): 13010–5. Bibcode:2006PNAS..10313010F. doi:10.1073/pnas.0603638103. JSTOR 30050705. PMC 1559744. PMID 16916934.
- ^ Maeda H, Dudareva N (2012). "The shikimate pathway and aromatic amino Acid biosynthesis in plants". Annual Review of Plant Biology. 63 (1): 73–105. doi:10.1146/annurev-arplant-042811-105439. PMID 22554242.
The AAA pathways consist of the shikimate pathway (the prechorismate pathway) and individual postchorismate pathways leading to Trp, Phe, and Tyr.... These pathways are found in bacteria, fungi, plants, and some protists but are absent in animals. Therefore, AAAs and some of their derivatives (vitamins) are essential nutrients in the human diet, although in animals Tyr can be synthesized from Phe by Phe hydroxylase....The absence of the AAA pathways in animals also makes these pathways attractive targets for antimicrobial agents and herbicides.
- ^ Cerdeira AL, Duke SO (2006). "The current status and environmental impacts of glyphosate-resistant crops: a review". Journal of Environmental Quality. 35 (5): 1633–58. doi:10.2134/jeq2005.0378. PMID 16899736.
- ^ "8.18.4.1.1. EPSP synthase: A tetrahedral ketal phosphate enzyme intermediate". Comprehensive Natural Products II. Chemistry and Biology. Reference Module in Chemistry, Molecular Sciences and Chemical Engineering. Vol. 8. 2010. pp. 663–688.
- ^ Anderson, Karen S.; Sammons, R. Douglas; Leo, Gregory C.; Sikorski, James A.; Benesi, Alan J.; Johnson, Kenneth A. (1990). "Observation by carbon-13 NMR of the EPSP synthase tetrahedral intermediate bound to the enzyme active site". Biochemistry. 29 (6): 1460–1465. doi:10.1021/bi00458a017. PMID 2334707.
- ^ Park, HaJeung; Hilsenbeck, Jacqueline L.; Kim, Hak Jun; Shuttleworth, Wendy A.; Park, Yong Ho; Evans, Jeremy N.; Kang, ChulHee (2004). "Structural studies of Streptococcus pneumoniae EPSP synthase in unliganded state, tetrahedral intermediate‐bound state and S3P‐GLP‐bound state". Molecular Microbiology. 51 (4): 963–971. doi:10.1046/j.1365-2958.2003.03885.x. PMID 14763973. S2CID 45549442.
- ^ Anderson, Karen S.; Sikorski, James A.; Johnson, Kenneth A. (1988). "A tetrahedral intermediate in the EPSP synthase reaction observed by rapid quench kinetics". Biochemistry. 27 (19): 7395–7406. doi:10.1021/bi00419a034. PMID 3061457.
- ^ Fonseca, Emily C. M.; da Costa, Kauê S.; Lameira, Jerônimo; Alves, Cláudio Nahum; Lima, Anderson H. (2020). "Investigation of the target-site resistance of EPSP synthase mutants P106T and T102I/P106S against glyphosate". RSC Advances. 10 (72): 44352–44360. doi:10.1039/D0RA09061A. ISSN 2046-2069. PMC 9058485.
- ^ Schönbrunn E, Eschenburg S, Shuttleworth WA, Schloss JV, Amrhein N, Evans JN, Kabsch W (Feb 2001). "Interaction of the herbicide glyphosate with its target enzyme 5-enolpyruvylshikimate 3-phosphate synthase in atomic detail". Proceedings of the National Academy of Sciences of the United States of America. 98 (4): 1376–80. Bibcode:2001PNAS...98.1376S. doi:10.1073/pnas.98.4.1376. PMC 29264. PMID 11171958.
- ^ Green JM, Owen MD (Jun 2011). "Herbicide-resistant crops: utilities and limitations for herbicide-resistant weed management". Journal of Agricultural and Food Chemistry. 59 (11): 5819–29. doi:10.1021/jf101286h. PMC 3105486. PMID 20586458.
Further reading
- Morell H, Clark MJ, Knowles PF, Sprinson DB (Jan 1967). "The enzymic synthesis of chorismic and prephenic acids from 3-enolpyruvylshikimic acid 5-phosphate". The Journal of Biological Chemistry. 242 (1): 82–90. doi:10.1016/S0021-9258(18)96321-0. PMID 4289188.